Exploring Soybean genomes
The most extensive capture of genetic diversity among cultivated accessions from a single country to date!
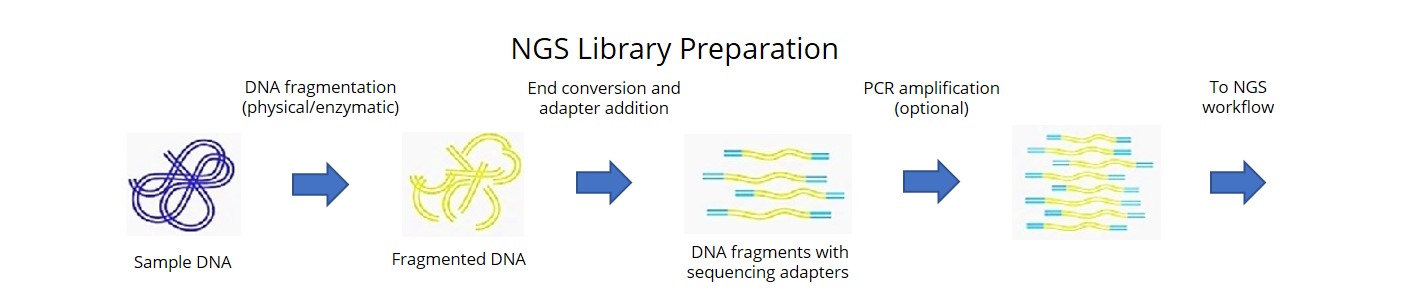
In this study, we sequenced a representative set of 102 short-season soybeans and achieved an extensive coverage of both nucleotide diversity and structural variation (SV).
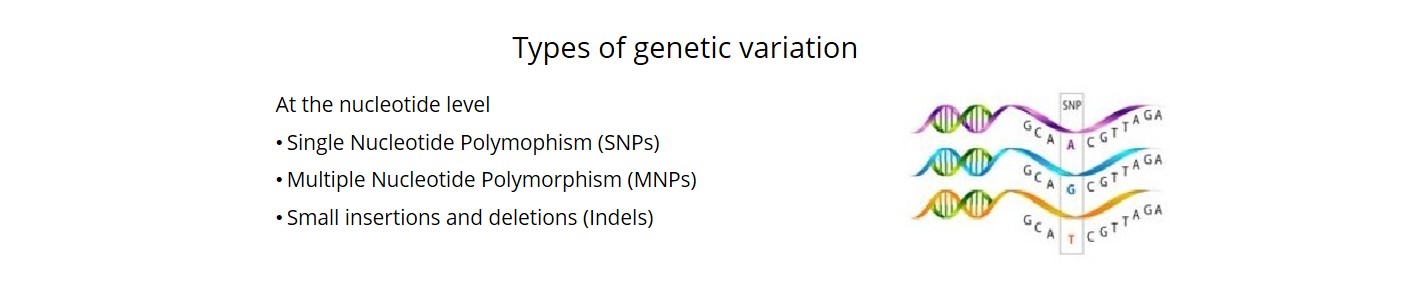
- We called close to 5M sequence variants (SNPs, MNPs and indels) with high level of accuracy (98.6%).
- We used this catalogue of SNPs as a reference panel to successfully impute (96.4%) missing genotypes at untyped loci in data sets derived from lower density genotyping tools (150 K GBS-derived SNPs/530 samples).
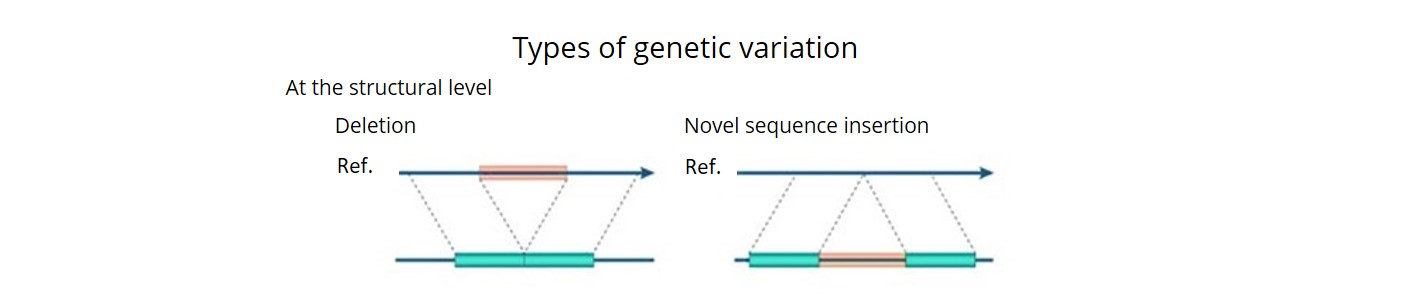
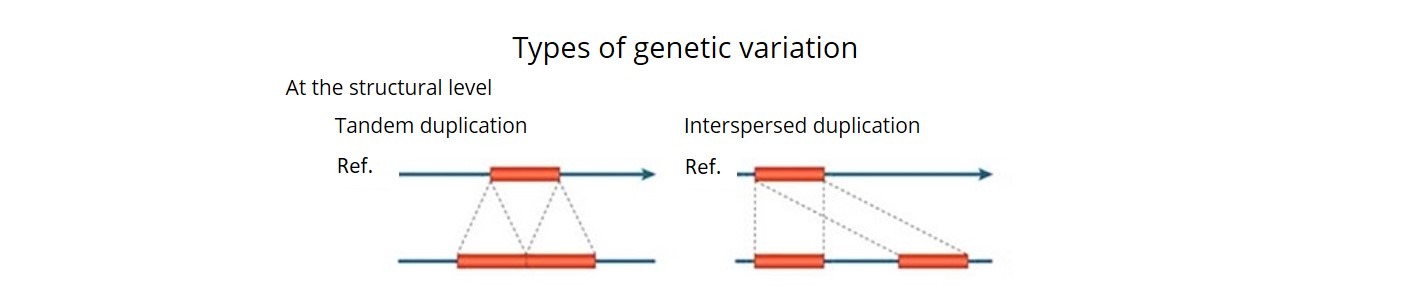
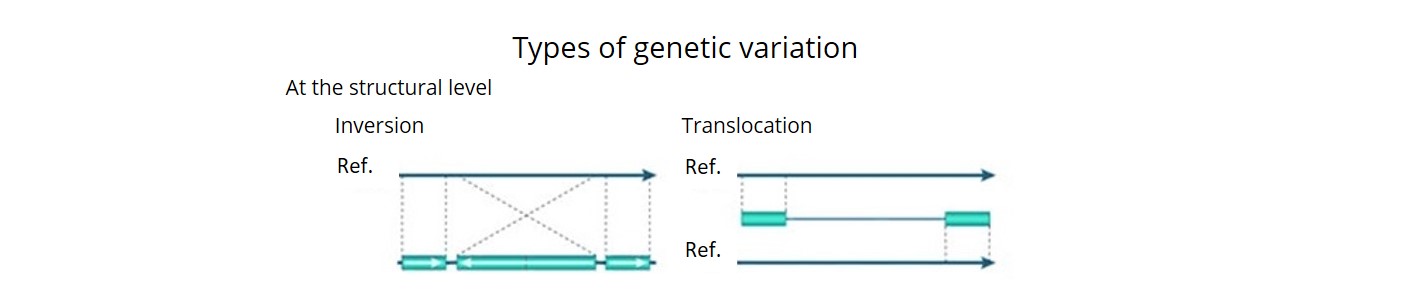
- Using a combination of three bioinformatics pipelines, we uncovered ~92 K SVs (deletions, insertions, inversions, duplications, CNVs and translocations) and estimated that over 90% of these were accurate. This is the first time that a comprehensive description of both SNP haplotype diversity and SV has been achieved within a regionally relevant subset of a major crop.